|
Algorithm introduction
Because this algorithm is difficult to understand, I use a example to illustrate it. Using base algorithm is KNN model(K-nearest neighbor algorithm). On the basis of the original model, we abstract four parameters to control the filling rate and accurating rate of imputing missing data and NPUTE program only have one parameters to setting. So our program can do better than NPUTE. Next, I will illustrate these four parameter and what's the meaning of the parameters. the four parameter is as follows:
-w, --windowSize
define window size
-k, --K-value
define the number of neighbor
-p, --noequalPunish
define punish when two genotypes are not same. noequalPunish must be integer. Normally gapScore is 5, equalScore is 10
-r, --percentRatio
define the ratio which the most genotype account for in all neighbor genotype
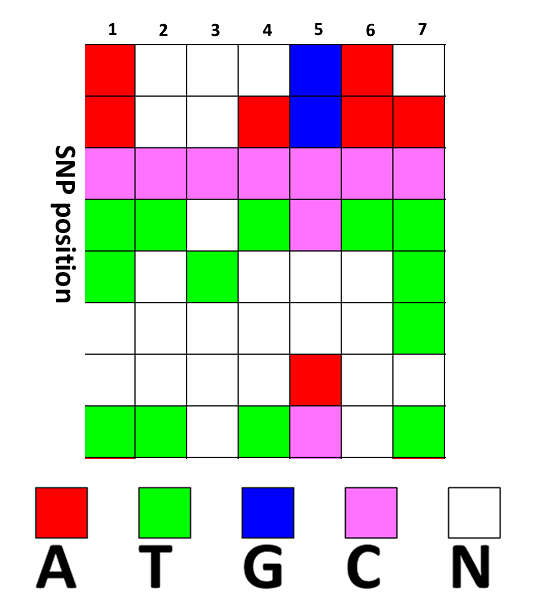
Input file format: the input file is M rows N columns matrix. ROW is SNP site,meanwhile COLUMN is inbreeding line. Missed genotype is represented by '-'. the below file is sample data which have 8 SNP sites and 7 inbreeding lines. For the cell, diffent color stands for different type of base pair, the cell white color will be imputed by our program.
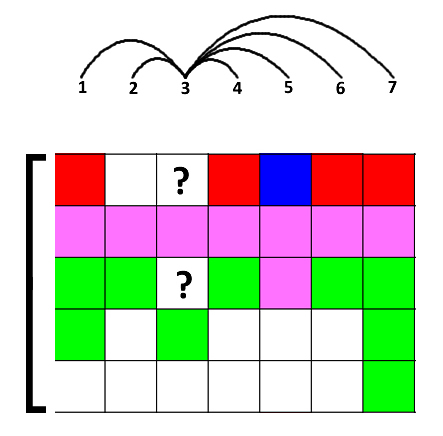
1. -w, --windowSize
Windows size simulates linkage disequilibrium(LD) in biology,
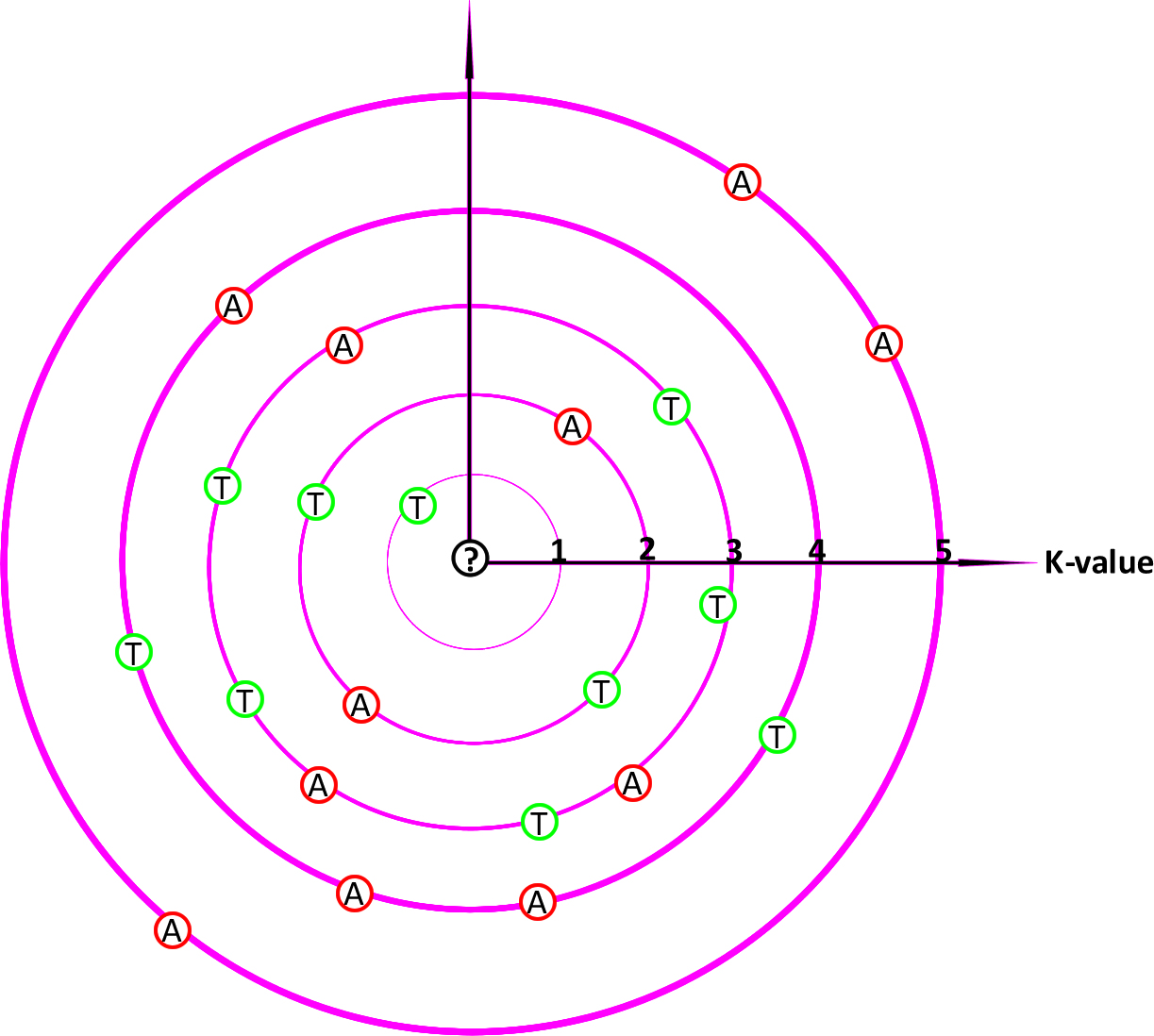
2. -k, --K-value
define the number of neighbor
3. -p, --noequalPunish
define punish when two genotypes are not same. noequalPunish must be integer. Normally gapScore is 5, equalScore is 10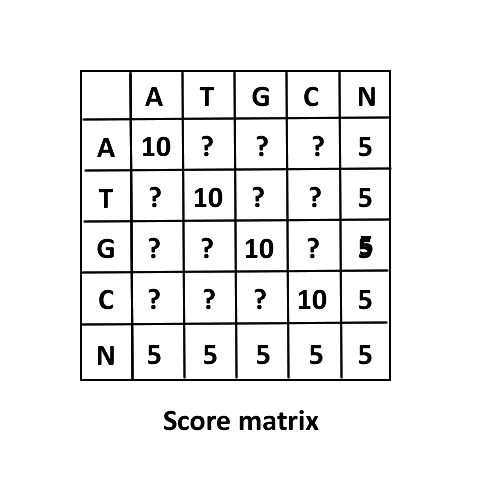
4. -r, --percentRatio
define the ratio which the most genotype account for in all neighbor genotype
After introducing the four parameters, we show the performace of this program comparing with NPUTE,fastPHASE. Morever we also call our fillGenotype program as Fimg (Filling missing genotype)
1. Fimg compares with NPUTE about costing time and using memory.
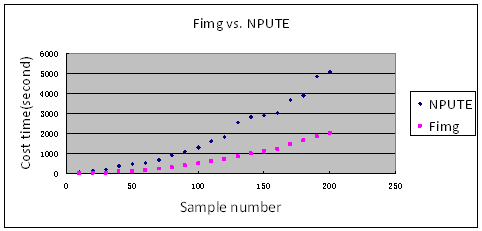
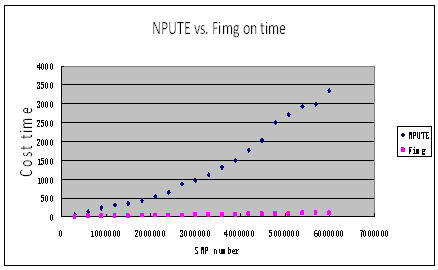
2. Fimg compares with NPUTE,fastPHASE about filling rate and accurating rate
|